Fast, detailed imaging across a wide field of view useful for deciphering brain functions
Fast, detailed imaging across a wide field of view useful for deciphering brain functions
WASHINGTON--(BUSINESS WIRE)-- Researchers have developed a microscope specifically for imaging large groups of interacting cells in their natural environments. The instrument provides scientists with a new tool for imaging neurons in living animals and could provide an unprecedented view into how large networks of neurons interact during various behaviors.
This press release features multimedia. View the full release here: https://www.businesswire.com/news/home/20190321005598/en/
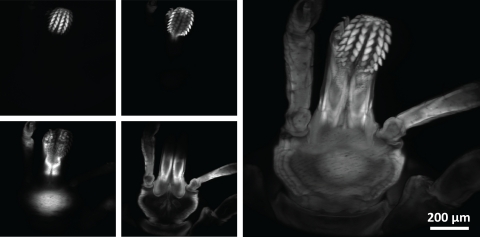
Caption: The researchers used the new microscope to image a tick. On the left are images from various depths, which were reconstructed into the volume image on the right. Credit: Amaury Badon, Boston University
In Optica, The Optical Society's journal for high-impact research, researchers from Boston University, USA show that their new “multi-z” confocal microscopy system can image the brains of living mice at video rate and with a field of view larger than a millimeter.
Imaging large groups of cells requires capturing cellular or subcellular details at fast speeds over a large 3D volume. This is challenging because most imaging approaches come with inherent tradeoffs between speed, field of view and resolution.
“We found a way to merge the needed imaging features in a microscopy system that is easy to build and operate,” said Amaury Badon, first author of the paper. “It also provides results in real time without the need for complicated data analysis or image processing.”
Acquiring 3D image volumes
The new microscope is based on confocal microscopy, a technique commonly used for cell imaging. Confocal microscopy produces images with high resolution and contrast by using a physical pinhole to block out-of-focus light and let in-focus light through. However, scanning a sample to acquire enough 2D images to reconstruct a 3D volume is time-consuming and produces large amounts of data.
To acquire multiple planes simultaneously, the researchers developed a way to reuse the light for imaging cells in one plane to also image cells deeper in the sample. They used an approach called extended illumination in which the microscope’s objective lens is only partially filled with the illuminating light, allowing the light to reach deeper into the sample. The full objective lens is then used to detect fluorescence, which provides high resolution. Rather than having one pinhole, like traditional confocal setups, the new microscope has a series of reflective pinholes that each capture in-focus light from a different depth within the sample.
“Our method benefits from the contrast of confocal microscopy while being able to extend to volumetric imaging without sacrificing speed,” said Badon. “Although extended illumination and reflective pinholes have been used before, this is the first time they were combined in a confocal microscope setup in a light-efficient way.”
The researchers also tailored the microscope for larger scale imaging than conventional confocal microscopes and designed it to image at video rate. Fast image acquisition was important because the fluorescence indicators that monitor cellular function typically operate on time scales of a few tens of milliseconds.
Imaging neural activity in live animals
The researchers demonstrated the multi-z confocal microscopy system by using it to image whole C. elegans worms, which are too large (500 to 800 microns long) to easily image all at once with a traditional confocal microscope. They simultaneously detected and monitored the activity of 42 neurons in the whole organism, even when the worms were moving.
They then used their microscope to image the hippocampal region of a mouse brain in an awake animal whose head was kept stationary. They were able to image neuron activity within a volume measuring 1200 X 1200 X 100 microns at video rate. Using an algorithm, the researchers were able to identify 926 neurons in the imaged volume.
They are now working to improve the speed and depth penetration of the technique as well as to make the microscope as versatile and user-friendly as possible.
Paper: A. Badon, S. Bensussen, H. J. Gritton, M. R. Awal, C. V. Gabel, X. Han, J. Mertz, “Video-rate large-scale imaging with multi-z confocal microscopy,” Optica, 6, 4, 389-395 (2019).
DOI: https://doi.org/10.1364/OPTICA.6.000389
About Optica
Optica is an open-access, online-only journal dedicated to the rapid dissemination of high-impact peer-reviewed research across the entire spectrum of optics and photonics. Published monthly by The Optical Society (OSA), Optica provides a forum for pioneering research to be swiftly accessed by the international community, whether that research is theoretical or experimental, fundamental or applied. Optica maintains a distinguished editorial board of more than 60 associate editors from around the world and is overseen by Editor-in-Chief Alex Gaeta, Columbia University, USA. For more information, visit Optica.
About The Optical Society
Founded in 1916, The Optical Society (OSA) is the leading professional organization for scientists, engineers, students and business leaders who fuel discoveries, shape real-life applications and accelerate achievements in the science of light. Through world-renowned publications, meetings and membership initiatives, OSA provides quality research, inspired interactions and dedicated resources for its extensive global network of optics and photonics experts. For more information, visit osa.org.
View source version on businesswire.com: https://www.businesswire.com/news/home/20190321005598/en/
Contacts
Media Contacts:
Aaron Cohen
(301) 633-6773
aaroncohenpr@gmail.com
mediarelations@osa.org
Source: The Optical Society
Smart Multimedia Gallery
Caption: The researchers used the new microscope to image a tick. On the left are images from various depths, which were reconstructed into the volume image on the right. Credit: Amaury Badon, Boston University