On August 21, the State Key Laboratory of Genome and Multi-omics Technologies, led by BGI-Research, collaborated with multiple institutions to unveil a groundbreaking single-cell omics technology named Stereo-cell in Science. This technology represents a transformative advancement in cellular analysis, promising to revolutionize precision medicine, rare disease research, and world’s fundamental understanding of biological systems.
Based on this technology platform, the Laboratory collaborates with 18 institutions to launch the “10 Billion Cells Alliance” (10BC). The alliance, which brings together global scientists to advance the decoding of the fundamental principles of life, will play a critical role in fostering open global collaboration and accelerating the translation of cutting-edge research into tangible public health benefits.
Wang Jian, Chairman and Co-founder of BGI Group, stated, "New technological breakthroughs will lead to new scientific discoveries. We hope to use this technology to serve more people, not only in China but also to make a greater impact internationally."
The study “Stereo-cell: Spatial
enhanced-resolution single-cell sequencing with high-density DNA
nanoball-patterned arrays” was published in Science.
Over
the past decade, single-cell sequencing technologies have dramatically advanced
global understanding of cellular heterogeneity and biological complexity,
enabling analysis of genomes, epigenomes, and transcriptomes at single-cell
resolution. However, existing single-cell approaches still face significant
challenges, including throughput limitations, capture uniformity issues, cell
size compatibility constraints, and technical scalability barriers. Published
in Science, Stereo-cell represents a breakthrough that overcomes these
long-standing limitations. Stereo-cell uses a chip densely tiled with DNA
nanoballs (DNBs) as "landing pads" for individual cells, serving as an array of nanoscale capture elements. These elements,
spaced at high density across the chip surface, enable precise spatial
positioning of individual cells.
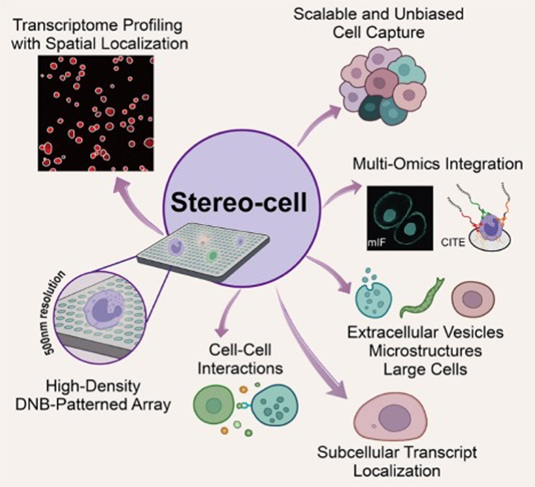
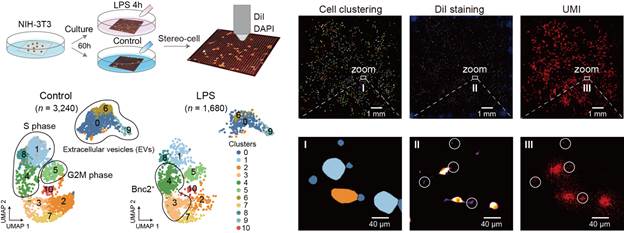
Stereo-cell on high-density DNB arrays
enables spatially resolved, multimodal single-cell profiling across scales;
this integrative design supports rare-cell discovery, microenvironment
analysis, and large-structure studies.
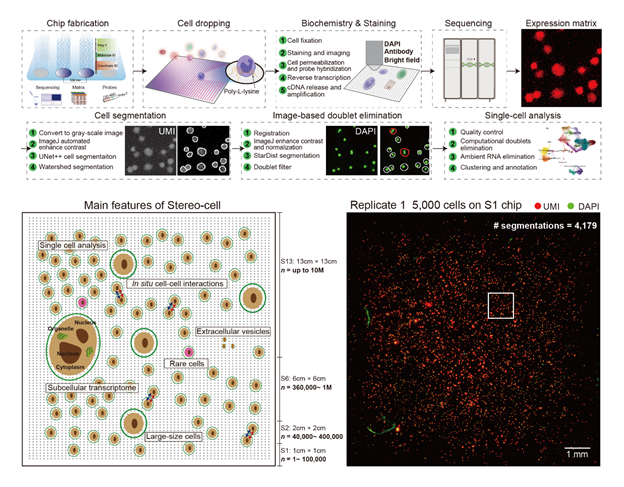
Cells
are deposited onto a poly-L-lysine–coated surface that enhances electrostatic
interactions to enable cell attachment, where RNA is captured by embedded
oligo-dT probes, imaging identifies cell positions, and sequencing reads transcripts.
This direct capture approach eliminates the need for droplet-based
encapsulation while enabling precise spatial positioning of individual cells
and maintaining their morphological integrity. Furthermore, optional workflows
enable multiplex immunofluorescence and protein profiling, while on-chip
culture allows time-resolved measurements of cellular dynamics. Up: Droplet-free, imaging-guided in situ
capture and deep-learning segmentation on a poly-L-lysine DNB array power
accurate single-cell calling; this lowers doublets and strengthens multimodal
readouts. Down: Scalable chips and spatial UMI
maps with thousands of segmentations demonstrate high-throughput, unbiased
capture; this scale enables detection of rare populations and robust atlas
construction.
The
study demonstrates strong performance across multiple datasets and cell types.
On a 6 by 6 cm chip, the team captured 445,467 peripheral blood cells in a
single experiment and detected rare hematopoietic stem and progenitor cells
(HSPCs) at approximately 0.05% of the population, achieving what researchers
describe as "finding needles in a haystack" at unprecedented scale. This
capability could prove crucial for early disease detection, as many diseases
begin with changes in rare cell populations that traditional methods might
miss. Imaging-guided filtering reduced doublets from 4.38% to 1.29% in mixed
human-mouse cell tests, demonstrating improved accuracy over traditional
droplet-based methods. In
benchmarks using human peripheral blood mononuclear cells (PBMCs), Stereo-cell
achieved cell-type proportions closer to flow-cytometry measurements than
public datasets from mainstream droplet platforms, with comparable gene
detection metrics. This means the technology provides more accurate
representations of what actually exists in patients' blood samples. For
large cells, the oocyte dataset included 719 cells with an average of 8,972
genes per cell, enabling high-throughput studies of fertility and reproductive
health that were previously limited to analyzing just a few cells at a time. "In
a single experiment, millions of cells can be captured, along with their
morphological, transcriptional, and protein characteristics, enabling deeper
analysis of cellular pathological states," said Liu Chuanyu, co-first
author of the paper and researcher at the BGI-Research. "Undoubtedly,
Stereo-cell is a milestone in the progression from single-cell omics to
clinical cell omics, with the potential to play a significant role in disease
mechanism research and clinical translation."
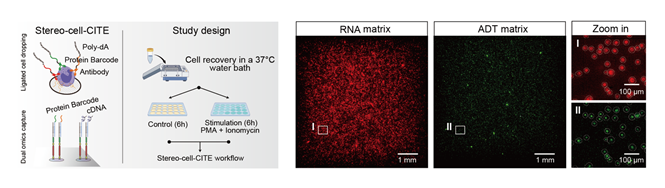
Left: Dual omics capture strategy of
Stereo-cell-CITE and study design. Right: Spatial visualization of the
distribution of captured RNA and protein on an S1 chip with an input of 10,000
human PBMCs.
The
authors describe multimodal advantages that provide insights beyond RNA
analysis alone, creating what researchers call "multidimensional cellular
profiles." Using multiplex immunofluorescence and Stereo-cell-CITE
workflows, protein markers including CD3, CD45RA, CD112-positive T cells, and
CD103-positive tissue-resident signatures matched transcriptomic clusters in
PBMCs, while stimulation experiments revealed regulatory networks in natural
killer cells. This work advances the world’s understanding of how immune cells
coordinate responses to threats. With
cells remaining in place on the chip, the team cultured fibroblasts directly on
arrays and recorded time-resolved changes, capturing elevated migration and
fibrosis pathway activity, providing new insights into wound healing and tissue
scarring processes. In multinucleated skeletal muscle fibers, Stereo-cell
defined spatial regions that localized gene modules at key junctions and
distinguished fiber-type markers, potentially informing treatments for muscle
diseases and age-related muscle loss.
Stereo-cell enables in situ sequencing
for cultured cells.
The
technology can also provide insights into cell–cell interactions,
microenvironments and subcellular localization under the study's experimental
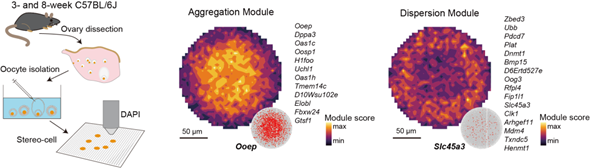
conditions. In oocytes, the authors mapped maturation trajectories and
identified RNA localization patterns consistent with single-molecule RNA FISH,
tracking the correlation between different subcellular
gene modules’ region-specific distributions within large cells. This knowledge
could advance fertility treatments and reproductive medicine.
Subcellular gene modules in oocytes
(e.g., OOEP aggregation, SLC45A3 dispersion) reveal organized RNA landscapes;
this resolution enables high-throughput mapping of intracellular regulation in
large cells.
Experts
have positioned Stereo-cell as a groundbreaking advancement in single-cell
analysis, transitioning from traditional "flat analysis" to
comprehensive "three-dimensional insights" that support large-scale
studies in cellular pathology, development, immune research, and genetics. They
emphasized four key advances: multimodal integration of spatial, RNA, and
protein signals; in situ, time-resolved readouts; compatibility with extreme
sample types; and scalability from hundreds to nearly a million cells per chip.
This breakthrough paves the way for transformative applications, including the
digitization of clinical pathology and large-scale drug screening. Professor
Wang Xiangdong, Chief Officer of Scientists at Zhongshan Hospital affiliated to
Fudan University, and Director of Shanghai Institute of Clinical Bioinformatics
and Fudan University Center of Clinical Bioinformatics, stated, "From a
clinical perspective, Stereo-cell technology has pioneered a new pathway in
clinical molecular medicine, which will help us provide better services for
patients." Currently, Professor Wang is collaborating with experts from
six hospitals, including Zhongshan Hospital of Fudan University, Shanghai
Tongji Hospital, and Henan Provincial People's Hospital, to form the
Stereo-cell clinical team. They are conducting projects on the clinical
translation of single-cell technology based on cutting-edge Stereo-cell
technology, aiming to provide patients with multidimensional and multi-faceted
clinical diagnostics and treatments. Professor
Ruan Yijun from the Life Sciences Institute at Zhejiang University stated:
"Stereo-cell is a groundbreaking technology. It has completely expanded
our imagination, enabling us to explore the functions of every human cell
during the life process, the changes that occur, and the conditions under which
diseases begin to develop. In the future, it will have unlimited application
prospects in the field of clinical medicine. I look forward to everyone working
together to advance single-cell technology from the billion-level to the
trillion-level, allowing us to truly and comprehensively decode the fate of
every cell in the human body." "Stereo-cell
is not just a technology platform, but a new generation of biological data
engine," said Xu Xun, co-corresponding author of the paper,
director of the State Key Laboratory of Genome and Multi-omics Technologies,
and chief scientist at BGI Group. "Based on this platform, the 10 Billion
Cells Alliance (10BC) was launched to construct the 'three major cellular
universe databases': life atlases, disease atlases, and perturbation-response
atlases. We welcome global research teams to collaborate and share, jointly
promoting the development of cell-scale AI foundation models and virtual cell
systems, and achieving a systematic leap from data to diagnosis and
treatment."
This
study can be accessed here: https://www.science.org/doi/10.1126/science.adr0475