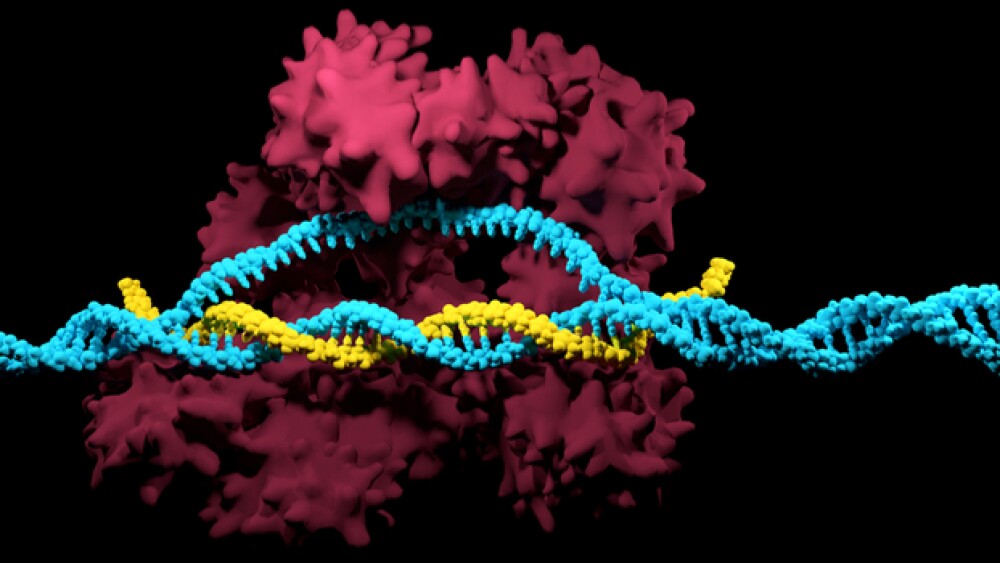
Yesterday, researchers at the Massachusetts Institute of Technology (MIT) published research in the journal Science Advances describing yet another CRISPR improvement.
When CRISPR was invented—or discovered—the gene editing system was expected to revolutionize research and medicine. And although it’s still early, the technique, which allows researchers to precisely and easily isolate gene sequences and replace them, has indeed shown promise. But researchers are also finding improvements that make the technique even more powerful.
For example, earlier this month, researchers at Children’s Hospital of Philadelphia (CHOP) used CRISPR to prevent a deadly metabolic disorder in prenatal laboratory animals. They used both CRISPR-Cas9 and base editor 3 (BE3) gene editing to cut cholesterol levels in healthy mice treated in utero. They targeted a gene that regulates those levels, and also used prenatal gene editing to improve liver function and prevent neonatal death in a subgroup of mice. The mice had been engineered with a mutation that caused the lethal liver disease hereditary tyrosinemia type 1 (HT1).
Although both early and conducted in lab animals, it shows promise for the potential of using CRISPR to treat some diseases prenatally.
Related, the CHOP researchers used a newer version of CRISPR called base editing, which was invented only two years ago. It changes an incorrect DNA letter—a base—to the correct or chosen one. CRISPR cuts DNA to do this, but base editing doesn’t. This potentially avoids unintended problems caused by DNA cuts.
Yesterday, researchers at the Massachusetts Institute of Technology (MIT) published research in the journal Science Advances describing yet another CRISPR improvement.
Traditional CRISPR using the Cas9 enzyme is limited in the number of locations in the genome it can hit. The reason is because CRISPR requires a specific sequence on either side of the target location, called a protospacer adjacent motif (PAM) in order to recognize the site.
An example is the most commonly used Cas9 enzyme, Streptococcus pyogenesCas9 (SpCas9), which needs two G nucleotides as its PAM sequence. That comes down to only about 9.9 percent of genome sites that can be targeted.
The MIT team, led by Joseph Jacobson, professor of media arts and sciences and head of the Molecular Machines research group, found a Cas9 enzyme that targets almost half of the genome locations.
“CRISPR is like a very accurate and efficient postal system, that can reach anywhere you want to go very precisely, but only if the ZIP code ends in a zero,” Jacobson stated. “So it is very accurate and specific, but it limits you greatly in the number of locations you can go to.”
The group developed a data analysis software tool they called SPAMALOT (Search for PAMs by Alignment of Targets), which searched bacterial sequences for similar enzymes with less restrictive PAM requirements.
They found several possibilities, but none jumped out as being great options. So they built synthetic versions in their laboratory and found the most successful was similar to the Cas9 enzyme already widely used.
“The enzyme looks almost identical to the one that was originally discovered … but it is able to target DNA sequences that the commonly used enzyme cannot,” stated Pranam Chatterjee, a graduate student in the Media Lab who conducted the research with fellow graduate student Noah Jakimo.
Instead of requiring two G nucleotides for its PAM sequence, it requires just one. This opens up the options for many more disease-specific mutations.
The research team is continuing to hunt for other enzymes with even broader powder. “We feel confident,” Jacobson stated, “of being able to go after every address on the genome.”